The origami of life
Imagine designing and mass-producing nano-sized objects that can interact with molecules and human cells with incredible precision. The structures would be precisely engineered to deliver drug therapies custom-fitted to each patient, or even bind to cancer cells to keep them from reproducing. Imagine being able to deploy a tiny device made of DNA that can detect the presence of a molecule that indicates a major health problem and release a drug.
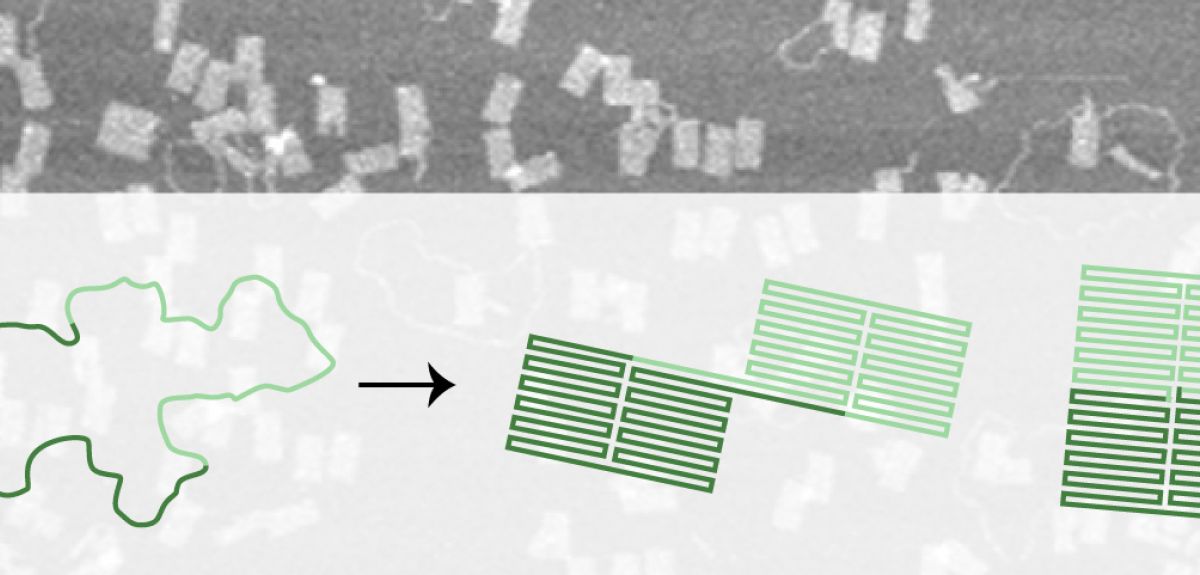
While these sorts of technologies are still a long way off, the process of designing them is advancing rapidly thanks to improvements in DNA origami technique. Nearly ten years ago, researchers first described a technique to fold a long single strand of DNA into various two- and three-dimensional shapes including a smiley face and three-dimensional cubes. The shapes are created by adding multiple small 'staple' strands of DNA to the template strand, gathering and doubling up the strand many times over as the mixture is combined, heated and then cooled in a buffer of salt water. Each drop of stapled DNA mixture contains around 1012 copies of each template strand and staples – if all goes to plan the result is 1012 identical DNA origami shapes. But it doesn’t always turn out that way.
'It is remarkable how well DNA origami works - in fact, it is remarkable that it works at all,' says Jonathan Bath, who is part of the biological physics group in Oxford’s Clarendon Laboratory. 'DNA origami is a reliable technique, if you design a simple structure you can expect a good fraction of your origami to be well-folded but as you increase the complexity of your design (for example when you go from two dimensions to three), the fraction of well-folded origami can fall dramatically.'
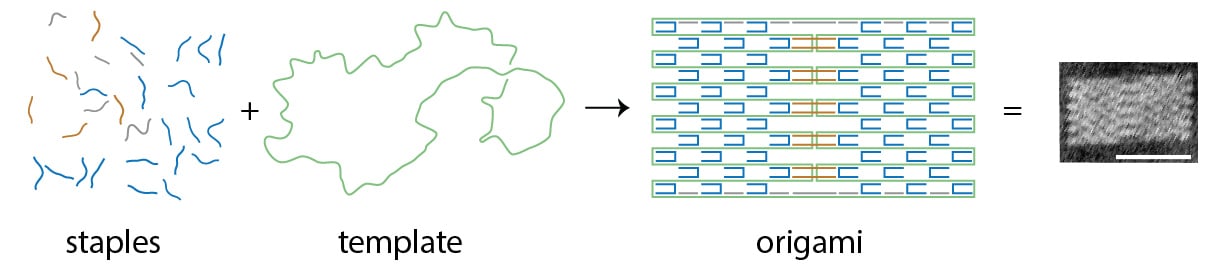
The way in which DNA strands and staples combine to form increasingly complex structures can be seen as both predictably obvious and confusing. DNA is a useful construction material because of the predictable way its base pairs join together. Its nucleotide bases Adenine (A), Guanine (G), Cytosine (C), and Thymine (T) form complementary A-T or C-G bonds, which means pairing staple DNA sequences in the right order to match a specific region of the template strand will produce predictable bonds that then produce shapes. Sections that are designed to bind together are given complementary sequences; other sections are given sequences that are as different as possible to minimize unintended interactions. What’s less obvious is how and why the stapled strands fold in sequence into well-formed shapes rather than just collapsing or mis-folding.
'Even the process of creating relatively simple DNA origami shapes produces a certain amount of mis-folded shapes,' Bath notes. 'As DNA origami structures get more sophisticated the technique becomes less reliable, and we end up with more mis-folded or tangled shapes that have to be separated out from the well-folded shapes we want to produce. If we want to fix this then we need to understand the folding. If we can fix it then we can continue to make increasingly sophisticated structures by guiding the folding pathways towards well-folded shapes and steer the system from becoming trapped in mis-folded shapes.'
This folding pathway is what Bath and his colleagues have investigated in a paper just published in Nature. Their goal, he emphasises, was not to make a 'new and improved' DNA origami structure but to design a system that would help to understand the mechanism that makes DNA origami fold in the way that it does. 'Often the first step towards understanding a system is to try and break it,' he explains, 'so we designed a DNA origami that has a handful of well folded-shapes but an overwhelmingly large number of misfolded states to trap the system.'
Remarkably, rather than breaking the DNA folding mechanism, Bath’s experiment still produced a surprising number of well-folded shapes. The result, he says, demonstrates the existence of folding pathways that steer the system towards well-folded shapes through a vast folding landscape littered with traps. ‘In any given experiment, we observe a mixture of well-folded shapes, some shapes more abundant than others,’ he explains. ‘The mixture serves as a record of events that take place along the folding pathway, it we tweak the folding pathway then the distribution of shapes that we observe will change.’
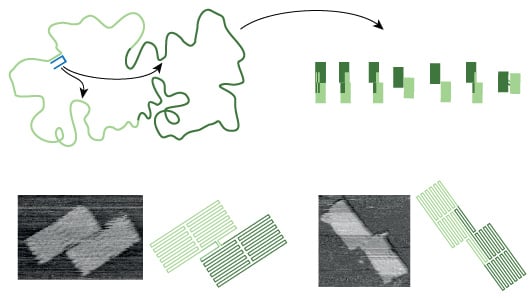
The experiment reveals that there are rules that govern the folding of DNA origami – if these rules are incorporated into future designs then they should make the production of complex DNA structures more successful. But the goal of the experiment wasn't just about improving DNA origami structures, but understanding them. 'Simple DNA origami folds well, so from a technology point of view, there is no problem,' says Bath. 'But from a science point of view there is a problem – we do not understand how it works and we would like to understand.'
'For me, the motivation comes from looking at biological systems: they are full of molecular machines –machines that read and copy information, join and break molecules, shuttle cargo from one place to another and so on. At least in principle, there is no reason why we should not be able to build our own machines to tackle similar tasks but this is a tall task and we’ll have to start simple. DNA nanotechnology (of which DNA origami is a subset) offers a realistic route towards the design and construction of molecular machines. If we are serious about achieving those ambitious goals then we need to understand what we are doing.'
The full report in the journal Nature is co-authored by Katherine Dunn, Jon Bath, Andrew Turberfield, Tom Ouldridge, Frits Dannenberg and Marta Kwiatkowska from Oxford University.