Oxford citizen science project helps improve detection of antibiotic resistance
Alison Farrar, a DPhil student researching how bacteria respond to antibiotic treatment at the Oxford Martin School, explains how citizen science and artificial intelligence are helping to combat the growing threat of antibiotic resistant bacteria.
Antibiotic resistance poses one of the most urgent challenges to public health worldwide. In this process, bacteria acquire genetic mutations that help them to become resistant to antibiotics. If bacteria become completely resistant to all antibiotics, this treatment will effectively become useless, and simple infections could cause deaths. In fact, this problem is already causing an estimated 1.3 million deaths every year.
One of the biggest challenges is that current testing methods can take up to two days to determine the most effective antibiotic for an infection. The goal of the Oxford Martin Programme on Antimicrobial Resistance Testing was to create a test that detects whether a patient’s bacteria are resistant to antibiotics within an hour. The test works by taking images of a patient’s bacteria under a microscope and using artificial intelligence (AI) to look for any changes that occur when antibiotics are applied to these samples. We recently published results from our citizen science project that investigated what makes some of these bacteria harder for AI to classify.
 Alison Farrar
Alison FarrarImage credit: Oxford Martin School
We collected thousands of images of resistant and sensitive bacteria treated with antibiotics. Bacteria that are sensitive to an antibiotic treatment develop changes to their shape, DNA, and cell wall. The AI model learns to detect these changes by studying images of bacteria that have responded to the antibiotic treatment and images of bacteria that don’t.
Sometimes, even though our AI model has seen thousands of images of antibiotic-treated bacteria, it still makes mistakes. In a recent research paper we showed that our current model is about 80% accurate at classifying each Escherichia coli (E. coli) cell. Although this leads to very high confidence when determining whether a whole sample is antibiotic sensitive or resistant, we want our diagnostic test to be as robust as possible.
We noticed that there was some variation in the extent to which E. coli cells changed after the antibiotic treatment, even when they were treated with the same concentration of antibiotic and had the same level of antibiotic resistance. In some cases, cells that looked like a resistant cell were actually sensitive, and vice versa. We started the Infection Inspection project to see which bacterial cells were most likely to be misinterpreted by volunteers, so that we could learn what features might also confuse the AI model. Then, we could focus on understanding those types of cells in our future research. We were also curious whether humans could detect more nuanced features than the AI model.
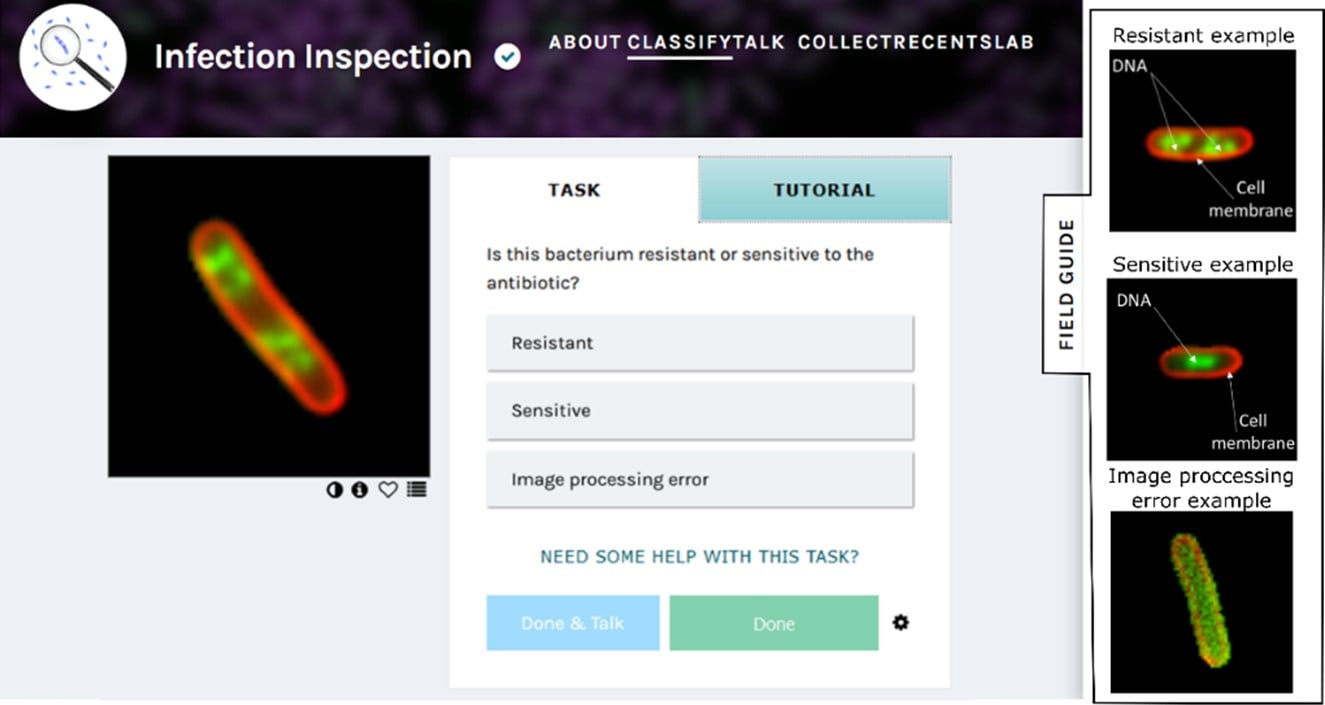
Volunteers were shown a picture of an E. coli cell that we treated with the antibiotic ciprofloxacin, stained, and imaged with our microscope. Because we grew the E. coli from strains collected at the hospital microbiology laboratory, we knew which strains were sensitive or resistant to ciprofloxacin on standard tests. Equipped with the field guide, volunteers could classify each image as resistant, sensitive, or an image processing error.
We were honoured that more than 5,000 volunteers contributed more than 1 million classifications to our project.
How difficult is it to classify a bacterial cell?
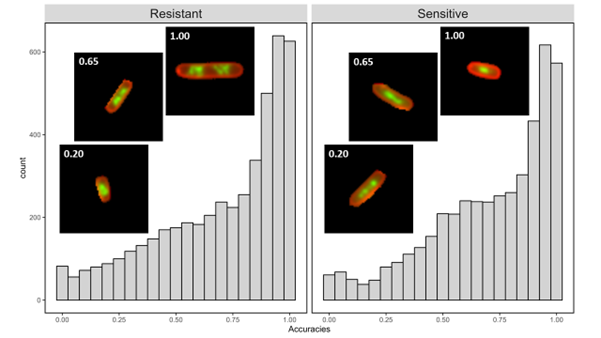
To understand which images were most likely to be misclassified as resistant or sensitive, we needed a way to measure whether a volunteer’s classification matched what we expected. We used E. coli cells from 5 clinical strains with different levels of resistance and treated them with different concentrations of ciprofloxacin for 30 minutes. We decided that if a cell was treated with an antibiotic concentration greater than its level of resistance, we expected it to look ‘sensitive’. In contrast, if a cell was treated with an antibiotic concentration less than its level of resistance, we’d expect it to look ‘resistant’. By defining these categories, we could discover when the biology didn’t match these predictions.
As we suspected, there was a lot of variety in how easy a cell was to classify and how well it lined up with our predictions. Some cells were classified correctly every single time, others most of the time, and others almost never. This was true for both ‘resistant’ and ‘sensitive’ cells. We could tell from images of cells that were rarely classified correctly that they had unusual or atypical DNA features.
From our analysis of the volunteer classifications, we couldn’t find any relationship between a volunteer’s accuracy and the number of images they classified or the number of days they were active on the project. It seemed like most of the difficulty of this task comes from the images themselves, rather than a user’s expertise.
What we learned and what comes next
The Infection Inspection project showed us that misclassifications of ciprofloxacin-sensitive and ciprofloxacin-resistant E. coli bacteria are associated with greater diversity in the appearance of the bacterial DNA after antibiotic treatment. It seems like most misclassifications happen when the features don’t line up with what we expect from a ‘sensitive’ or ‘resistant’ cell, rather than our AI misidentifying features.
Even though we expect the bacteria in each of our samples to be genetically identical, there are clearly some cells that respond differently than others. This is an area with lots of open questions that we are designing experiments to answer.
Some volunteers started to notice that in some of our images, it looked like a cell was in the process of cell reproduction. This idea could be related to why some of the bacteria in our samples respond differently than others. It’s possible that the stage of the bacterial life cycle at which the cell is exposed to the antibiotic has an impact on the appearance of the DNA. This is a question we’ll continue to explore in our research.
Thank you
Our group is actively looking into the antibiotic response of many bacterial species to different antibiotics, so that we can develop a rapid test for antibiotic resistance. There is a lot to learn here, and we are extremely grateful to the Zooniverse volunteers who participated. Their enthusiasm and curiosity were extraordinary, so thank you for your dedication and engagement. We’d also like to thank the Zooniverse platform leaders, Helen Spiers, Mary Westwood, and Cliff Johnson, for their expertise and contributions to the development of this project.
 25-year Oxford study finds the effects of conflict last for generations
25-year Oxford study finds the effects of conflict last for generations What Louise Thompson’s campaign tells us about the national maternity crisis
What Louise Thompson’s campaign tells us about the national maternity crisis Celebrating 25 Years of Clarendon
Celebrating 25 Years of Clarendon  Learning for peace: global governance education at Oxford
Learning for peace: global governance education at Oxford  What US intervention could mean for displaced Venezuelans
What US intervention could mean for displaced Venezuelans  10 years on: The Oxford learning centre making an impact
10 years on: The Oxford learning centre making an impact Oxford and The Brilliant Club: inspiring the next generation of scholars
Oxford and The Brilliant Club: inspiring the next generation of scholars New course launched for the next generation of creative translators
New course launched for the next generation of creative translators