Credit: Dr Peter Canning
Inflammatory remarks
Ponatinib is an anti-cancer drug which has earned some notoriety for its cost (£90,000 per patient per year) and side-effects that were serious enough to temporarily suspend its use. But the findings from a recent Cell Chemistry and Biology paper led by the Structural Genomics Consortium suggest that this 'dirty' drug might actually hold the key for coming up with newer, more effective drugs for chronic illnesses such as Crohn's and Inflammatory Bowel Disease.
The Oxford Science Blog asked the first author, Dr Peter Canning (who worked on this study while at the Nuffield Department of Medicine) to explain what they found.
OxSciBlog: Why were you interested in ponatinib?
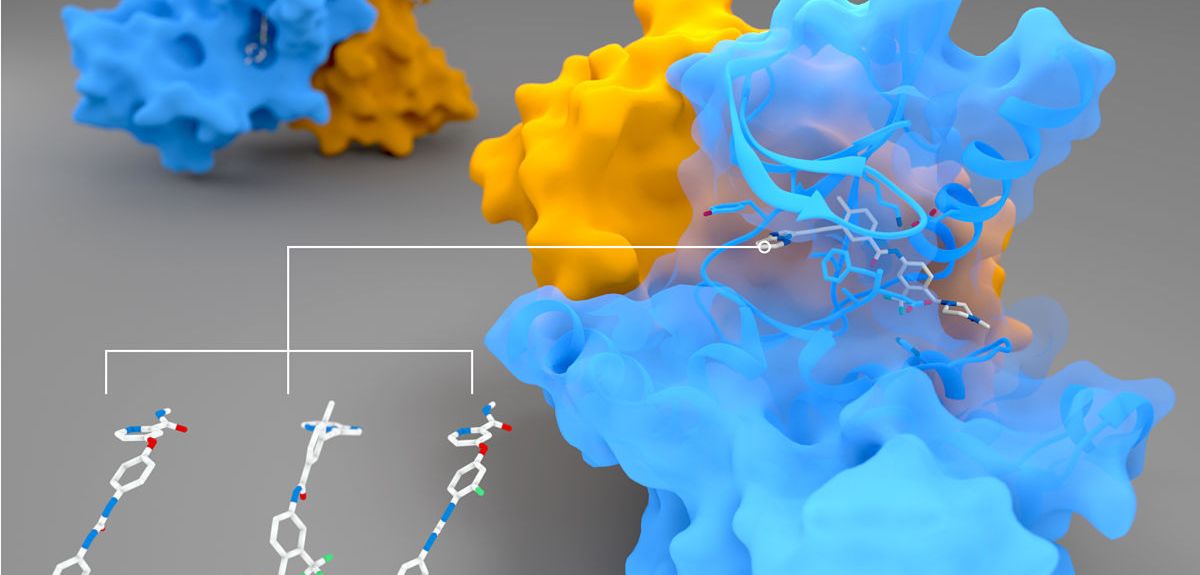
Peter Canning: Our main interest was originally in a protein called RIPK2 (pictured above), which has an important role in regulating the body's immune response. When the body is invaded by rogue bacteria, they can be detected by two human proteins, NOD1 and NOD2. But NOD1 and 2 need RIPK2 to pass on a signal that will activate an immune response to fight off the invasion.
This signalling can go wrong in autoimmune and inflammatory diseases, such as inflammatory bowel disease, and abnormal inflammatory signalling can even be a factor in some kinds of cancer. One potential cause of disease is if the NOD proteins start to signal constantly, even when there are no bacteria around. This then produces a constant signal from RIPK2, which in turn triggers a constant inflammatory response. If we can find ways of inhibiting this signal, we can stop it, and thus treat the disease. So RIPK2 is potentially a very interesting drug target for inflammatory diseases.
OSB: How do you go about finding inhibitors for a protein?
PC: We can use a number of methods, but one of the first screens we try at the Structural Genomics Consortium is to perform a 'thermal shift' experiment: we take the protein of interest, and we heat it.
Proteins are folded into a three dimensional shape, but when they get too hot, they unfold and lose their shape.
Proteins are folded into a three dimensional shape, but when they get too hot, they unfold and lose their shape. We exploit this change by using a dye that sticks to the unfolded protein core, and so it can only do so when the core is exposed. The dye is fluorescent, and you see these beautiful peaks in the fluorescent signal when the protein unfolds.
When we add a protein inhibitor into the mix, it has a stabilizing effect on the protein, so that it unfolds at a higher temperature. So you get an increase in the temperature at which you see a peak in the fluorescent signal.
OSB: So what happened when you added ponatinib to RIPK2 in this experiment?
PC: I’ve used this method to look for protein inhibitors many times, and the results with ponatinib were the most striking that I have ever seen. In my experience working on protein kinases like RIPK2, a good inhibitor might increase a protein’s unfolding temperature by about 10 ˚C.
However, with ponatinib, the melting point of RIPK2 increased by 23 ˚C. So that was immediately interesting.
OSB: Can you tell us a bit more about ponatinib, and how and why it was originally developed?
PC: Ponatinib is based on a drug called imatinib, which I've worked on before. Imatinib inhibits the BCR-ABL protein that causes chronic myelogenous leukaemia (a cancer of white blood cells) and gastrointestinal stromal tumours.
Imatinib was an amazing breakthrough that featured on the cover of Time magazine. It's a striking example of what is called target-based drug design
Imatinib was an amazing breakthrough that featured on the cover of Time magazine. It's a striking example of what is called target-based drug design: some clever scientists worked out that the specific fusion of two genes was producing the BCR-ABL fusion protein, and that this fusion protein was then resulting in the development of specific kinds of cancer. So they reasoned that targeting this specific protein would be a great treatment for this kind of cancer. And they turned out to be 100% right!
The problem is that to stop the cancer from coming back, you have to keep taking imatinib for the rest of your life. More or less inevitably, the disease-causing BCR-ABL eventually mutates, developing resistance to imatinib. So imatinib derivatives have now been designed to treat particular kinds of cancer when imatinib has stopped working.
One of these derivatives is ponatinib, which is designed to treat a very specific mutation of BCR-ABL. Its approval was actually retracted for a while, because it causes an increase in blood clots, leading to a greater risk of heart attacks and strokes in the patients taking it. It was then reapproved, but with restrictions on dosage levels and the conditions it can be used for: it’s really only for a subset of late-stage cancers.
OSB: How did you use ponatinib in your experiments?
PC: We wanted to solve the 3D structure of the RIPK2 protein to understand more about how it works as well as how drug molecules might bind. This involves using X-ray crystallography to determine the exact 3D arrangement of the atoms that make up the RIPK2 protein.
This is often a difficult challenge, as we first need to induce the protein to form into solid crystals. RIPK2 is a protein kinase, and it can be tricky to induce kinases to form crystals, due to their flexibility. However, if you add an inhibitor so that the protein locks into a particular 3D shape, it can make the protein a lot easier to crystallize.
Once we have grown a protein crystal, we take it to the Diamond Light Source facility, which produces incredibly powerful and fine beams of X-rays. We shine these powerful X-ray beams onto the protein, and that gives us a diffraction pattern of X-rays as they go through the crystal. Then we use computational methods to turn the diffraction pattern back into the structure of the thing that scattered the X-ray in the first place: this kind of X-ray crystallography was used to decode the structure of DNA as well. RIPK2 crystals were particular thin and small, and we were really only able to get our data because the X-rays at the Diamond Light Source are so incredibly focused.
But having ponatinib is what allowed us to stabilise RIPK2 enough for it to crystallize in the first place.
OSB: Why is it important to understand the structure of RIPK2, or any other protein?
PC: Understanding RIPK2's crystal structure not only helps us understand how protein kinases work, but structural information is incredibly useful if you're looking for a drug target. If, for example, we know that an inhibitor locks onto RIPK2 but also several other proteins, we can look for clues in the structure of the proteins to come up with a more specific inhibitor that will affect RIPK2 only, but not others: that is how you make good drugs that have low side-effects.
Understanding RIPK2's crystal structure not only helps us understand how protein kinases work, but structural information is incredibly useful if you're looking for a drug target
OSB: What other experiments did you do?
PC: We tried putting ponatinib on isolated human cells to see if it could block their inflammatory response by inhibiting RIPK2. And it turned out that it could.
We also looked at whether other similar drugs might also bind to RIPK2. As we hoped, we found three others that seemed to bind equally well. But when we put these drugs onto cells, we were surprised that only two produced the same potent effect as ponatinib. We don't fully know why that would happen.
One clue was that all the drugs (including ponatinib) that were very effective in cells were so-called 'type II kinase inhibitors': they work by binding to kinases and twisting them out of shape, thus blocking their function. The drug that didn't work as well was a 'type I' inhibitor: it simply blocks up the place where a key molecule (ATP) binds, and this may explain the difference. But we don't know for sure – it's a curious puzzle that we are still investigating!
OSB: Are these drugs potential treatments for inflammatory diseases then?
PC: The most marked effect in our experiments was from ponatinib, but it’s pretty unlikely that ponatinib would make for a good treatment. Its side-effects are too severe: inflammatory diseases like inflammatory bowel disease and Crohn's disease are chronic conditions. You’d need to take any treatment for a long time, and so you can't really risk something like increased heart attacks!
But thanks to ponatinib, we know the 3D structure of RIPK2, including its drug binding 'pocket', and this has provided us with some clues for how we might make modified drug molecules so that they target RIPK2 more selectively. With luck, such molecules could potentially provide a treatment for chronic inflammatory diseases in future.
Dr Peter Canning blogs at: https://petecanning.wordpress.com/